SARS-CoV-2 nucleoprotein from S. cerevisiae, frozen stock
Description: SARS-CoV-2 recombinant Nucleoprotein (SCoV2-rN) expressed in yeast Saccharomyces cerevisiae. UniProtKB sequence accession number: P0DTC9 (NCAP_SARS2), expressed with C-terminal His6-tag. It was purified from yeast lysates under native (non-denaturing) conditions using convenient chromatographic techniques. Then it was dialyzed against PBS, adjusted to 1.0 mg/ml concentration, filter-sterilized, aliquoted and frozen for storage.
Form: Frozen liquid in PBS (pH 7.4).
Safety: Product 20-S2N-ScA-F contains only yeast expressed proteins that are not infectious or hazard. Contains no plasma or serum of animal origin. No safety precautions are required.
Storage: The product is shipped with dry ice (shipment cost to Europe: 160 €). Store at -80°C for the long term. After thawing may be stored for short-term at 4°C (stable at least 1 week at this temperature). Avoid repeated freeze/thaw cycles.
Concentration: 1.0 mg/ml.
Image: SDS-PAGE showing SCoV2-rN product 20-S2N-ScA-F at approximately 49 kDa (3 μg/lane).
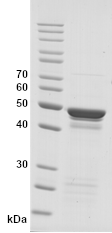
Purity: >85% pure intact form of SCoV2-rN protein as determined by SDS-PAGE. Most other protein bands represent minor partial degradation of the same protein.
Applications (tested): ELISA (IgG), other immunoassays, Total Internal Reflection Ellipsometry (TIRE), Electron microscopy, Western blot, SDS-PAGE.
Reactivity of SCoV2-rN protein (product 20-S2N-ScA-F) with specific IgG in ELISA using Covid-19 positive and negative sera: this product has been compared both with other commercial SARS-CoV-2 recombinant proteins (produced by other producers) in ELISA and S1 antigen-based CE-approved commercial IVD tests using the same set of human sera. Results for sensitivity and specificity are provided in the Table below:
|
Reference test for Covid-19 positive sera |
RT-PCR |
|||||
| Analyzed antigens or serological tests |
SARS-rN* IgG (Baltymas) |
SCoV2-rN (from E.coli) IgG (GeneTex) |
SCoV2-rN (# 20-S2N-ScA-F) IgG (Baltymas) | SARS2 S1 IgG (Sino Biological) | AMP rapid (S1) IgG test | Euroimmun ELISA (S1) IgG test |
| Sensitivity (n=50), correct positives | 92.00% | 92.00% | 92.00% | 78.00% | 90.00% | 90.00% |
| Specificity (n=150), correct negatives | 99.33% | 94.00% | 99.33% | 97.33% | 99.33% | 98.67% |
| OD mean for positive sera (n=50) | 1,967 | 1,455 | 2,502 | 0,980 | N. A. | 1,820 |
*SARS coronavirus recombinant protein (SARS-rN) was produced by Baltymas for comparison with homologue SARS-CoV-2 nucleoprotein. It has been shown that, due to high similarity of these proteins, SARS-CoV nucleoprotein may also be used for the detection of specific antibodies against SARS-CoV-2. Still, SCoV2-rN showed higher reactivity with specific antibodies in Covid-19 positive sera.
Interaction kinetics measured between specific polyclonal mouse antibodies against SARS-CoV2 N protein (anti-SCoV2-rN) and SCoV2-rN (product 20-S2N-ScA-F) by the total internal reflection ellipsometry (TIRE) method: association and dissociation rate constants, and association (affinity) and dissociation constants, as well as steric factor were calculated using appropriate mathematical models (Ref. 1). The calculations of steric factor indicated that SCoV2-rN/anti-SCoV2-rN immune complex formation has very strict orientation requirements (Ref. 1). These findings can be applied in the design of new antibody/antigen interaction-based analytical systems dedicated for the determination of anti-SCoV2-N antibodies and for the development of new anti-SARS-CoV-2 medications based on proteins that are blocking viral SCoV2-N proteins.
Unit/Price:
| Size | Catalogue No. | Price | |
| 100 μg | 20-S2N-ScA-F-C | 175 € | Inquiry / Order product |
| 500 μg | 20-S2N-ScA-F-D | 625 € | |
| 1 mg | 20-S2N-ScA-F-M | 1000 € |
Reference:
- Plikusiene I, Maciulis V, Ramanaviciene A, Balevicius Z, Buzavaite-Verteliene E, Ciplys E, Slibinskas R, Simanavicius M, Zvirbliene A, Ramanavicius A: Evaluation of kinetics and thermodynamics of interaction between immobilized SARS-CoV-2 nucleoprotein and specific antibodies by total internal reflection ellipsometry. J Colloid Interface Sci. 2021 Jul 15; 594:195-203. doi: 10.1016/j.jcis.2021.02.100. Full text.